Modeling Microenvironment Trajectories on Spatial Transcriptomics with NicheFlow
Abstract
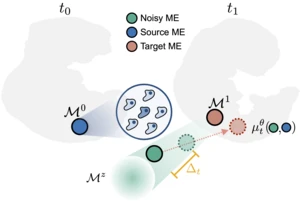
Understanding the evolution of cellular microenvironments is essential for deciphering tissue development and disease progression. While spatial transcriptomics now enables high-resolution mapping of tissue organization across space and time, current techniques that analyze cellular evolution operate at the single-cell level, overlooking critical spatial relationships. We introduce NicheFlow, a flow-based generative model that infers the temporal trajectory of cellular microenvironments across sequential spatial slides. By representing local cell neighborhoods as point clouds, NicheFlow jointly models the evolution of cell states and coordinates using optimal transport and Variational Flow Matching. Our approach successfully recovers both global spatial architecture and local microenvironment composition across diverse spatio-temporal datasets, from embryonic to brain development.
Links
Citation
Modeling Microenvironment Trajectories on Spatial Transcriptomics with NicheFlow
Kristiyan Sakalyan*, Alessandro Palma*, Filippo Guerranti*, Fabian J Theis, Stephan Günnemann
NeurIPS, 2025
If you use this work in your research, please cite this paper:
@inproceedings{sakalayan2025nicheflow,
title={Modeling Microenvironment Trajectories on Spatial Transcriptomics with NicheFlow},
author={Sakalayan, Kristiyan and Palma, Alessandro and Guerranti, Filippo and Theis, Fabian and G{\"u}nnemann, Stephan},
booktitle={Neural Information Processing Systems (NeurIPS)},
year={2025},
url={https://openreview.net/forum?id=5ofJyjgrth}
}